Abstract
Background: Mantle cell lymphoma (MCL) is a non-Hodgkin lymphoma that is incurable. MCL has a complex ecosystem of malignant B-cells and stromal and immune cells that play a supporting role for tumor growth, ultimately leading to the potential re-emergence of the disease. The tumor microenvironment has been reported as a crucial factor in MCL pathogenesis and progression. Thus, if we can identify the tumor microenvironment components and define the characteristics of malignant and non-malignant cells, this will pave the way for studying clonal evolution of MCL in vivo.
Methods: Both pre- and post-treatment, fresh tumor biopsy samples of MCL were obtained. The cells were dissociated and re-suspended in PBS with >10% serum. A final concentration of 1,200 cells/uL were used for single cell sorting in the chromium system (10X Genomics, California). We sequenced the mRNA in the NextSeq 500 platform. All analysis was conducted using R-programming language (version 3.4).
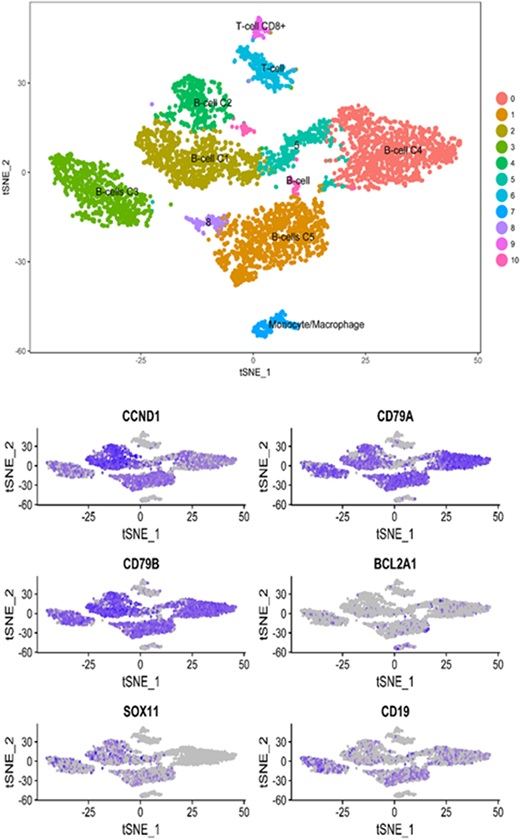
Results: From four MCL patients (L1-L4), we obtained 9,400 cells. Three of the four samples were collected through apheresis (i.e., L1-L3), and one sample (i.e., L4) from surgical biopsy of the involved lymph node. One patient had known TP53 mutated status (i.e., L1) and another patient had CCND1 translocation (i.e., L2). From the apheresis samples (L1-L3), the proportion of lymphocytes was 87%, 68% and 65%. We identified 10 defined clusters of cells based upon their gene expression from all four samples. Six of the 10 clusters were clonal B-cells with strong expression of CCND1, CD79A and CD79B. We also identified clonal T-cells (both CD8+ and CD4+) and monocyte/macrophage clusters. SOX11 expression was absent in one B-cell clone, indicating this clone may be SOX11-negative MCL. The monocyte-macrophage cluster demonstrated strong BCL2 expression, which was not expressed by the B-cells clones. CD19 expression was ubiquitous among the B-cell clones but weaker as compared with other B-cell markers. When the signaling was compared among the four samples, the chemo-resistant cells (sample L1) demonstrated upregulation of NOTCH1 signaling, DNA-damage repair, interferon-alpha response, MYC targets and the HIF1A pathway. Two cell clones did not express any canonical markers and were not identified as a defined cluster.
Conclusions: We identified meaningful sub-populations of MCL that define the tumor microenvironment. There is considerable inter-tumor and intra-tumor heterogeneity of MCL at a single cell resolution, which indicates that developing a uniform treatment regimen may prove to be difficult. Ubiquitous expression of CD79A or CD79B may help guiding precision medicine such as the development of novel CAR-T cell therapy. Longitudinal follow up of the same patients may define clonal evolution of MCL and unravel the spatio-temporal interplay.
Wang:AstraZeneca: Consultancy, Research Funding; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Kite Pharma: Research Funding; Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; MoreHealth: Consultancy; Novartis: Research Funding; Acerta Pharma: Honoraria, Research Funding; Dava Oncology: Honoraria; Juno: Research Funding; Pharmacyclics: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal